Diving Deep In To Sourvisiae
Last week I posted a deep-dive into the fermentation and re-pitchablity of the Philly Sour strain of Lachancea thermotolerans. In parallel with those experiments I was also testing the characteristics of Sourvisiae. Sourvisiae is a genetically engineered sour fermenting yeast from Lallemand. It is intended to make quick sours without needing to add bacteria. It is also advertised (and has a reputation of being) a very clean fermenter. In this post I’m going to go into how this yeast was made, how it works, and provide some information on how to best use – and re-use – this yeast in your brewery.
Note: Figures 3 and 6 had their colour schemes reversed in an earlier version of this article. They have since been corrected.
Click this link if you want to jump straight to the tests.
A General Note/Warning
This yeast is a GMO. Unfortunately, a number of “environmental” NGOs and organic food producers have spent a lot of money trying to convince you that GMO’s are dangerous.
In reality, GMOs are some of the most heavily scrutinized products in the history of human kind, and have been consumed by humans and animals for over three decades. There are literally thousands of safety (and environmental) studies on GMO’s showing them to be safe.
I will not tolerate any fearmongering about GMOs in the comments. This is not what this post is about. If you want to debate GMO safety, do so elsewhere. Comments violating this policy will be deleted; repeat offenders will be blocked from commenting. My blog, my rules.
How Was Sourvisiae Made – The Big View
As part of the approvals process, Mascoma (the division of Lallemand whom made Sourvisiae) had to reveal how they made Sourvisiae This means that we can look quite closely at how this yeast was engineered. I’m not going to go over every detail of their approval, but the full documentation can be found here.
To start, almost all eukaryotes (the domain of life which includes fungi, yeast, plants, and all animals including yourself) are capable of generating lactic acid – this includes conventional brewers yeast. This reaction is fairly simple, and uses much of the same biochemical pathway as does alcohol fermentation:

So if brewers yeast has the capability of making lactic acid, why doesn’t it?
Simply put, it comes down to evolution. Brewers yeast expresses (makes) a lot of the enzymes which produce alcohol, but only small amounts of the enzyme which makes lactic acid. In addition, the ethanol-generating enzymes in yeast are very fast and efficient, while the lactic acid generating enzyme is not. The net effect is that only small amounts of lactic acid are made during conventional yeast fermentation.
If you look at the metabolic diagram (Figure 1) you’ll notice that it only takes the enzyme lactate dehydrogenase to make lactic acid. Meaning it’s theoretically simple to make a lactic acid producing yeast. All you need to do is give it a highly expressed copy of an efficient lactate dehydrogenase.
And that is what Mascoma did.
How Was Sourvisiae Made – The Fine Details
Of course, how the actual yeast was made is a bit more complex than what I described above. While you can often just take a gene from one organism and drop it into another, doing so often leads to inefficient gene expression or expression when you don’t want it. As such, a few steps are usually taken to ensure that the gene is expressed at the right level, and at the right time. But before we go into that, we briefly need to mention how genes work (Figure 2).

- Engineering the promotor: The promotor is what controls when a gene is expressed, and how strongly it is expressed. So when moving genes between organisms you usually replace the genes natural promotor with a promotor from the recipient organism. You can select a promotor that is active when you want it to be active, and at the desired level.
- Optimizing the gene: The genetic code is universal. This means that you can move a gene from one organism to the next without modification and the resulting protein will be the same. But the way organisms use the code can differ. DNA is read in 3-letter “blocks”, with each block (called a codon) coding for a single amino acid in the resulting protein. Some amino acids are coded for by more than one codon. For example, the amino acid glycine can be coded in DNA as ‘GGT’, ‘GGA’, ‘GGG’ and ‘GGC’. Different species will “prefer” to use one or two of those codons over the others. As such, we often optimize codons to use those preferred by the recipient organism. Without this step, you may not get as much protein made as you expect.
- Selecting a terminator: The end of the gene is the terminator. This part is not incorporated into the protein, but it can have an effect on how long the mRNA lasts – and therefore how much protein gets made from each mRNA.
The folks at Mascoma did all three of these steps:
- Promotor: The promotor from the yeast alcohol dehydrogenase 1 (ADH1) gene – e.g. the gene used to make alcohol from acetaldehyde – was used. This ensures that the new gene is active any time the yeast is performing fermentation.
- Gene: The lactate dehydrogenase gene (LDH1) from the fungus Rhizopus oryzae was codon optimized for yeast, and the optimized version added after the ADH1 promotor. R. oryzae is commonly found in the soil and on our food, and is considered safe for human exposure and consumption. It has a very active and efficient lactate dehydrogenase gene.
- Terminator: The terminator from the yeast’s pyruvate decarboxylase 1 (PDC1) was used. PDC1 is one of the genes needed for glycolysis.
These three elements were cloned together and then inserted into the yeast’s genome using a method called “recombination”. Unlike many other methods of inserting genes into genomes, this method allows you to insert your gene with no extra genetic material into a desired location in the recipients cell’s genome. Thus, the newly generated yeast contain only the engineered gene and no other new genetic features. This is also a very stable way to alter the cell, meaning that the added gene should not be “lost” as the yeast reproduces.
The exact yeast strain used to make Sourvisiae is, to my knowledge, not known outside of Mascoma/Lallemand. That said, I strongly suspect it is either US-05 or another closely related Chico strain.
Characterization
To keep my results as consistent as possible, all of the tests unless otherwise noted were performed using hop-free wort made from dry malt extract, at a gravity of 1.045. All fermentations were performed in 50 mL volumes at 200C. All wort was sterilized in the mini-fermenters, using an Instant Pot, high pressure setting for 15 minutes (and yes, this truly sterilized the wort). Unless otherwise noted, the manufacturers minimum recommended pitch rate of 2.5 million yeast were pitched per millilitre of wort. Yeast densities were confirmed by hemocytometer, using trypan blue to ensure only live yeast were counted.
Attenuation was generally compelted within 5 days. However, all ferments were conducted for 10 days to ensure consistency with the Philly Sour experiments.
General Fermentation Characteristics
I started off with the same battery of tests that I began my Philly Sour experiments with – fermentation dynamics, pitch rate versus terminal pH, and pH versus original gravity. Sourvisiae is a faster fermenter than Philly Sour, and also acidifies evenly during the fermentation process (Figure 3). Both terminal gravity and terminal pH were reached by day 5 of fermentation in my tests.

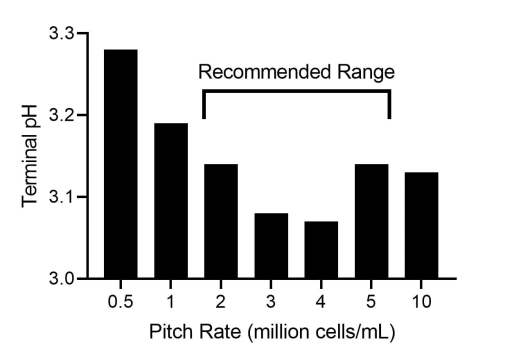
The pitching rate also had a much weaker effect on terminal pH than I observed with Philly Sour (Figure 4). Only severe under-pitching lead to reduced acidification. The under-pitched worts also attenuated poorly (F.G. of 1.020 instead of the usual 1.011-1.013, not shown).
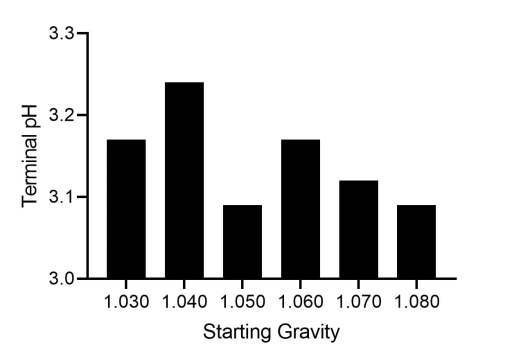
Sourvisiae produced a similar terminal pH across a broad range of starting gravities (Figure 5).
Repitching & Other Issues
One common comment made about Sourvisiae is that it appears to produce less acid upon repitch, leading some to speculate that the genetic modification is lost over time. This is likely untrue, as in their approval Moscoma shows that the genetic modification is stable for 100 generations. In comparison, on average, yeast divide 3 or so times during a beer fermentation. So I repitched the yeast, every time repitiching 2.5 million viable yeast per mL of media (Figure 6). The same terminal pH was reached after each repitch. So where does this claim come from?

Due to the intense sourness of this yeast, it is often co-pitched with another strain. This could explain this result, if Sourvisiae divided more slowly, or died more quickly post-ferment, than other yeasts. So I performed a bear of an experiment, where I performed hourly cells counts covering the 0-24 hour growth period. This was performed with cultures of Sourvisiae, Nottingham, W34-70, and US-05 (Figure 7). Sourvisiae has a generation time that is 20% to 30% longer than any of these yeasts (Table 1).

| Yeast | Generation Time (hrs) |
|---|---|
| US-05 | 3.27 |
| Nottingham | 3.58 |
| W34-70 | 3.63 |
| Sourvisiae | 4.62 |
I also looked at the death of Sourvisiae following fermentation, compared to US-05 in fermented wort adjusted to the same terminal pH (3.2) using lactic acid. Unlike Philly Sour, Sourvisiae did not die more quickly than US-05 under these conditions (Figure 8). The slower growth may explain the “lost” acidification power reported by brewers, as in a mixed culture, the proportion of Sourvisiae cells will continually decrease.

This also opens up the possibility that Sourvisiae could easily be overwhelmed by even a modest contamination. Mathematically, given the ~30% faster growth of US-05 over Sourvisiae, even a million-to-1 ratio of Sourvisiae:US-05 would lead to a measurable (1%) amount of US-05 by the 9th repitch (Figure 9). To test this possibility I cast agar plates with 0.1% (w/v) chalk incorporated into the media. When plated on this medium, yeast which produce a lot of acid will neutralize the chalk in the surrounding gel, leaving a clear area around the colony. Conventional brewers yeast do not produce sufficient acidity to produce a clear area. I plan on posting a video on this method sometime in 2021…so more on this method will be posted on a later date.

I then started a test fermentation with a million-to-1 mixture of Sourvisiae to US-05 (serial dilutions are a wonderful thing). Each fermentation was allowed to go for 10 days before repitching into the next fermenter, with each repitch at 2.5 million viable cells per mL. I began plating the yeast on the chalk-wort medium starting on generation 8, which according to my model should be the generation prior to a detectable population of US-05. This should occur at ~1% contamination, as I’m counting plates with ~200 colonies. Surprisingly, acid-negative yeasts (e.g. US-05) were already ~1% of the population by this time-point. US-05 was the predominant cell type by generation 12 (Figure 10).

Final Thoughts
Sourvisiae has some advantages over Philly Sour. Consistent acidification is much less dependent on pitch rate, and you don’t need to use tricks like adding glucose to get good acidification. It ferments faster, allowing beer to be completed sooner. It also survives better post-fermentation, giving brewers more flexibility in re-pitching the yeast.
Some aspects of Sourvisiae are more of a draw compared to Philly Sour. On a pH scale the difference in terminal pH between Philly Sour (3.4-3.5) and Sourvisiae (3.1-3.2) don’t seem all that different. But pH is a logarithmic scale. This means that there is between 1.75 and 5 times as much acidity in a Sourvisiae ferment. This is notable when you taste these samples – Philly Sour is typically pleasantly tart, while Sourvisiae boarders on undrinkably sour.
There are some definite disadvantages as well. The slower growth means that Sourvisiae can be overwhelmed by a contaminating yeast or when used in a blend, which limits re-pitching. Sourvisiae is also a rather bland yeast, and in my opinion, Philly Sour gives a finish that is more complementary to dry-hoped and fruited sours.
I have an upcoming post (or maybe video, I haven’t decided yet) comparing the two in the same base recipe…so more on flavour and other characteristics will be posted at a later date.




This is great, thanks for sharing your work.
In the experiment where you tracked cell counts for the 4 strains over 24 hours, I’d be interested to hear more about the fermentation setup. Were these also 50mL fermentations, with samples taken continuously from single flasks (per strain) over the time period? Or were you destroying samples after taking a cell count (which would really be a bear of a setup)? Were these static or agitated fermentations?
Mostly I’m curious about how you overcame the issue of obtaining a representative sample in active fermentation. I know from experience that can be tricky depending on your fermentation size and vessel.
The same 50 mL fermentations as used throughout the post. These are 125 mL Erlenmeyer flasks with 50 mL of 1.040 wort (made from DME, no hops). As I am trying to mimic conventional fermentation, no stirring or shaking was used (outside of sampling). In all cases, a known (counted by hemocytometer) amount of yeast was pitched to each flask, from an active and stirred starter.
As I stated in the post, the growth measurements were taken from two flasks per yeast strain – one flask inoculated 12 hours before “time 0” and the other inoculated at “time 0”. Every hour I gently swirled the flasks to even out the yeast, and then used a micropipette to pull a sample which was counted on a hemocytometer. I did cell counts with a hemocytometer from each flask every hour, over 12 hours. This way, the first flask (the -12hr flask) gave me measurements for hours 12 to 24 of the growth curve, and the second flask gave me measurements for hours 0-11 of the growth curve. All counts were done in duplicate (e.g. separate samples were pulled at each time point, and loaded onto opposite sides of the hemocytometer), with the graph shows the average value of those duplicates.
That makes a lot of sense, I appreciate the details! This is a bit divergent from the topic of this particular post, but do you think you would see any difference in overall cell growth over that 24 hour period if samples could be taken without agitation? Or by destroying samples as they’re pulled?
At the pitch rate I was using I don’t think any differences would be seen after 24 hours – all the cultures had reached stationary phase, and you typically don’t see further division at that point. I don’t think the sampling had any effect on the results. The volume I needed for the individual time-points (0.02 mL) was 0.04% of the total fermentation volume, meaning that a total of 0.48% of the total volume (and yeast) were “consumed” by the testing,
I just finished a brew of 10 gallons of American wheat beer at an O.G. of 1.041 with Sourvisiae and ended up with a three-day ferment that ended at 1.012 (3.8% ABV) and a pH I estimate at 3.5 or so (my pH strips bottomed out at 4.0 so this is a guess). The final beer was super clean, but also super sour and very one-dimensional. I put 5 gallons on 2# of tart plums and the other 5 gallons on ginger and bitter orange peel. The ginger and orange is definitely the better batch, as the tart plum simply enhanced the already sharp tartness. I am considering blending a regular non-hopped American wheat into the beers to soften them and make them more approachable. I am also considering using a split pitch, for my next brew, of regular saccharomyces with the Sourvisciae to see if they can work side-by-side to produce a more approachable sour. Any thoughts on blending yeasts?
I’m trying some blended yeasts right now, and I’ve shared sourvisiae with a few members of my brew club as well who’ve tried blends. If co-pitching, it looks like somewhere between 25% sourvisiae to 50% sourvisiae gives a decent sourness that isn’t overpowering. One of my club members tried about 10% sourvisiae and didn’t get noticeable sourness. Make sure the yeast you are co-pitching is strong and healthy, as it will likely struggle as the pH drops.
Blending with another beer also works well. I blended the dry-hoped sourvisiae beer with an American IPA (~2:1 sourvisiae:IPA) and the resulting blend was fantastic – slightly tart, great hop character.
Very interesting! Have you thought about looking at the metabolic flux of sugars/pyruvate to EtOH vs. Lactic acid for Sourvisiae?
Also, I wonder if the slower growth of sourvisiae vs US 05 is related to stress caused by the production of high concentrations of lactic acid that must be transported out of the cell? It would be interesting to look at organic acid transport gene expression in Sourvisiae vs. US 05 to see if there is increased expression, although not directly relevant to most brewers!
I don’t have the technology in my home lab to look at pyruvate or other metabolic aspects. I’d be willing to bet though that the slower growth is a duirect product of the stresses put on the cells by the lactic acid production – especially as the modifications introduced into the yeast did not include an extra lactate transporter to help the cells get rid of the lactic acid.
Well done testing of two souring agents. Thanks and look forward to the comparison. Would also be nice to titrate % added US 05 or K97 against Sourvisiae to see where pH and TA wind up. ie… what is best ratio for a good sour but no overly sour Gose or Berliner
I’ve seen a few co-pitching rates out there; I think lallemand suggests 10% US-05 or nottingham as a good place to start.
I’m brewing the comparison this weekend (weather permitting), so hopefully I’ll have the comparison in a month.